By Rebecca Schmidt, Ana Cristina Fulladolsa Palma and Vamsi Nalam (9/23)

Quick Facts
- Accurate diagnosis of plant diseases is essential for disease management in agriculture.
- Proper sampling of plant tissues is crucial for accurate disease diagnosis.
- Molecular diagnostic methods significantly improve accuracy but have practical considerations, such as time and trained personnel.
- Standard molecular methods like ELISA and PCR must be performed in the lab, while others, such as the lateral flow assay and RPA, can be performed outside of a lab.
Why is plant diagnostics essential?
Accurate and rapid diagnosis of plant diseases is essential for effective disease management in agriculture. Early detection of pathogens (microorganisms that cause disease) allows farmers to implement control measures before diseases spread and cause significant crop damage. Accurate pathogen identification informs the selection of disease-resistant plant varieties in the future and appropriate pesticide use, leading to more effective disease management strategies. Incorrect diagnoses can result in considerable economic losses for farmers.
Traditional methods of plant disease identification, such as microscopy and culture-based assays, can be time-consuming and require specialized equipment and trained personnel. These issues can limit the availability of accurate diagnoses, especially in remote or resource-limited areas.
Advances in molecular biology have led to the development of diagnostic tools such as ELISA (enzyme-linked immunosorbent assay), lateral flow assay, PCR (polymerase chain reaction), RPA(recombinase polymerase amplification), and next-generation sequencing. These tools provide rapid and accurate plant disease diagnoses that help farmers make informed decisions and minimize the impact of plant diseases on their crops.
Effective Sampling
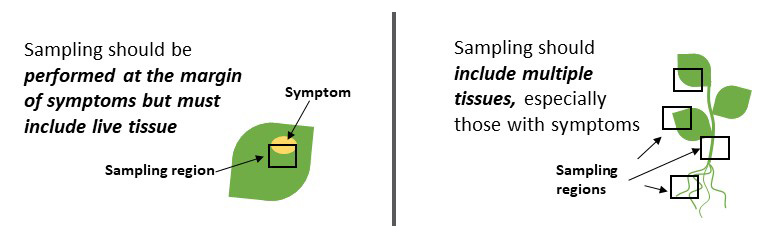
Collecting good plant samples for disease diagnosis is important to ensure accurate diagnosis. Sampling from living tissue that is symptomatic is essential. Sampling from diverse tissues such as root, stem, and leaves is helpful when the pathogen causing the symptoms is unknown (Figure 1).
Pathogens can live in specific tissues or move among plant tissues throughout their life cycle, so access to various tissues increases the likelihood of finding the cause of the problem. For more detailed information on sample collection, please refer to Fact Sheet 7.246 and the CSU Plant Diagnostic clinic (https://agsci.colostate.edu/agbio/plantclinic).
Figure 1. Plant sampling considerations. Effective sampling should include living tissue from symptomatic areas, and include a variety of tissues. The black boxes represent sampling areas.
Methods for Molecular Diagnosis
1. ELISA (Enzyme-linked Immunosorbent Assay)
ELISA detects and measures levels of an antigen, specific pathogen proteins, in a sample (Fig. 2). It uses a specific antibody that can bind to the protein of interest, and an enzyme is used to detect this binding. Antibodies used in ELISA are highly specific and can accurately differentiate between similar pathogens. Sample preparation for this method is minimal and is performed by grinding plant tissue in an extraction buffer. Though ELISA is in expensive and suitable for numerous samples, it may be time-intensive. The presence of a pathogen is usually indicated by a color change, which is simple to interpret but can be subtle and may require specialized equipment and personnel trained to interpret results.
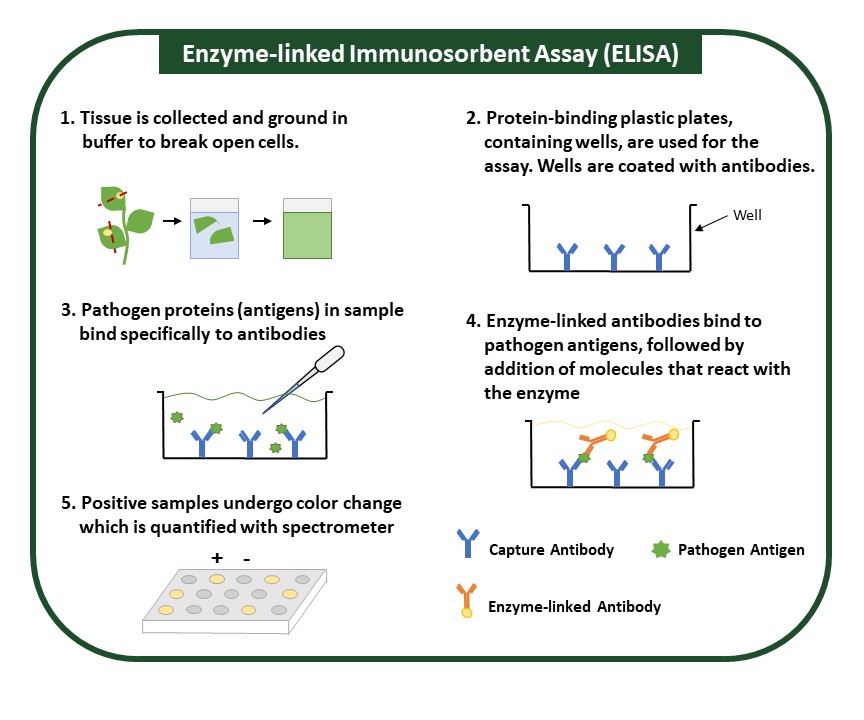
Figure 2. Enzyme-linked Immunosorbent Assay (ELISA). ELISA uses pathogen-targeted antibodies to identify pathogens within a sample. A positive result, or presence of a pathogen in a sample, is indicated by a color change that can be quantified with a spectrometer.
2. Lateral Flow Assay
The method uses antibodies to identify specific pathogen proteins (antigens). Sample preparation for this method is minimal and can be performed by grinding plant tissue in an extraction buffer. The sample is absorbed along the test strip where, if present, pathogen antigens first bind to labeled antibodies. As they continue to move along the test strip, pathogen antigens will bind to a second antibody at the test line, resulting in a positive test line (Figure 3).
Antibodies used in this method are highly specific and can accurately differentiate between similar pathogens. However, this method is more costly than ELISA and cannot easily be altered to accommodate multiple samples. Nevertheless, the assay is simple, and results can be obtained in under 30 minutes. Results can be interpreted visually on the test strip, and all steps can be performed at room temperature, which means this assay can be performed in the field. Lateral flow assays can also target pathogen nucleic acids (DNA or RNA) and are designed similarly to those described in this section.
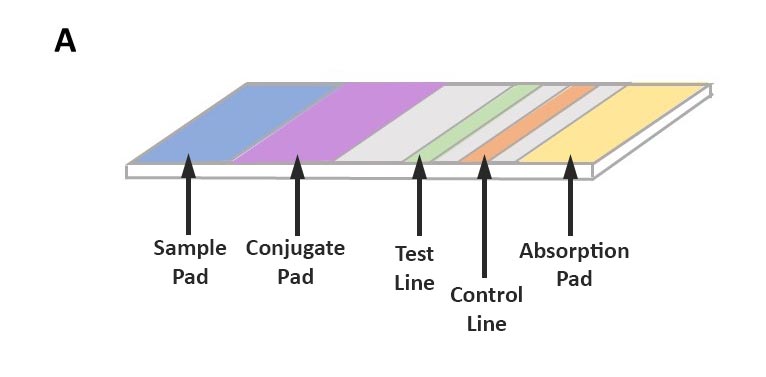
Figure 3A: Structure of lateral flow assay test strip. The sample is absorbed into the test strip at the sample pad and passes through each section, the conjugate pad, the test line, and the control line, finally ending at the absorption pad. The test strip is usually made of a nitrocellulose membrane.
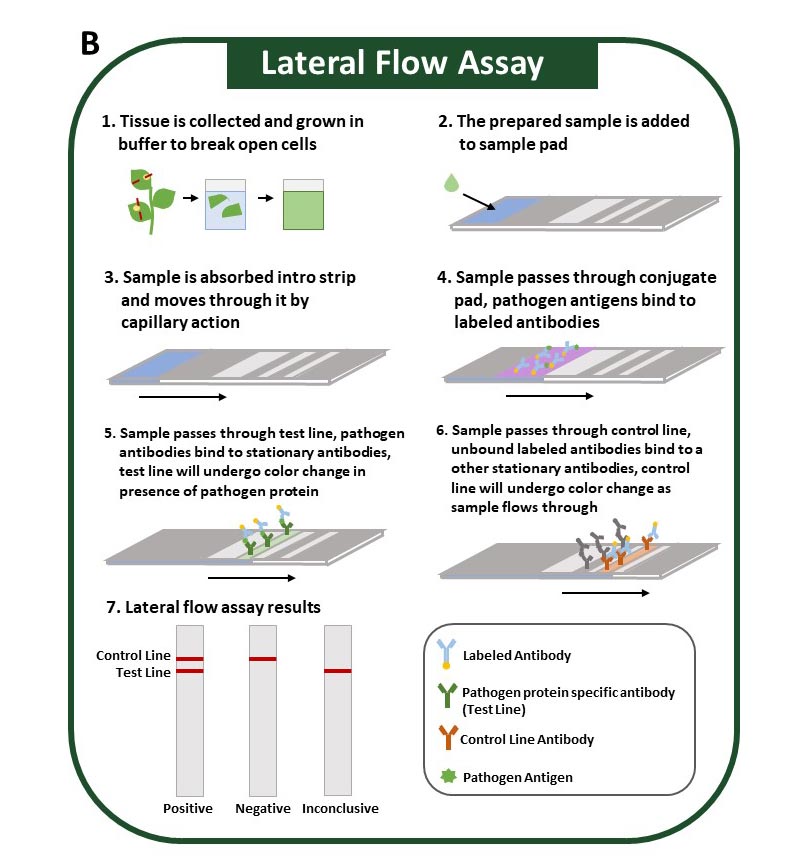
Figure 3B: Lateral flow assay uses pathogen-targeted antibodies to identify pathogens within a sample. Positive tests are determined visually on the test strip.
PCR (Polymerase Chain Reaction)
PCR utilizes an enzyme to amplify specific regions of pathogen genomes, which allows a pathogen in the sample to be identified (Figure 4). The PCR reaction is carried out in a thermocycler, which allows each step of amplification, denaturation, annealing, and elongation to occur at optimal temperatures (Figure 4). Samples that contain the pathogen will show DNA amplification, while samples that do not have the pathogen will not.
A PCR reaction contains several components: (1) Primers, which target a specific part of the pathogen genome, (2) polymerase, an enzyme that elongates the DNA, (3) dNTPs, nucleotides that are used by enzymes; and (4) salts, which provide optimal conditions for the reaction. Because primers can be designed for any pathogen whose genomic sequences are known, PCR can identify various pathogens, from bacteria to viruses. However, it may be difficult to distinguish between different pathogen strains due to their high degree of genomic similarity.
PCR is more sensitive because target DNA from pathogens is amplified, allowing pathogen detection at low infection levels. Sample preparation for this method requires extraction of DNA or RNA; therefore, it must be performed in a laboratory. In addition, performing the PCR, or amplification reaction, requires samples to be incubated at multiple temperatures and therefore requires equipment and some expertise.
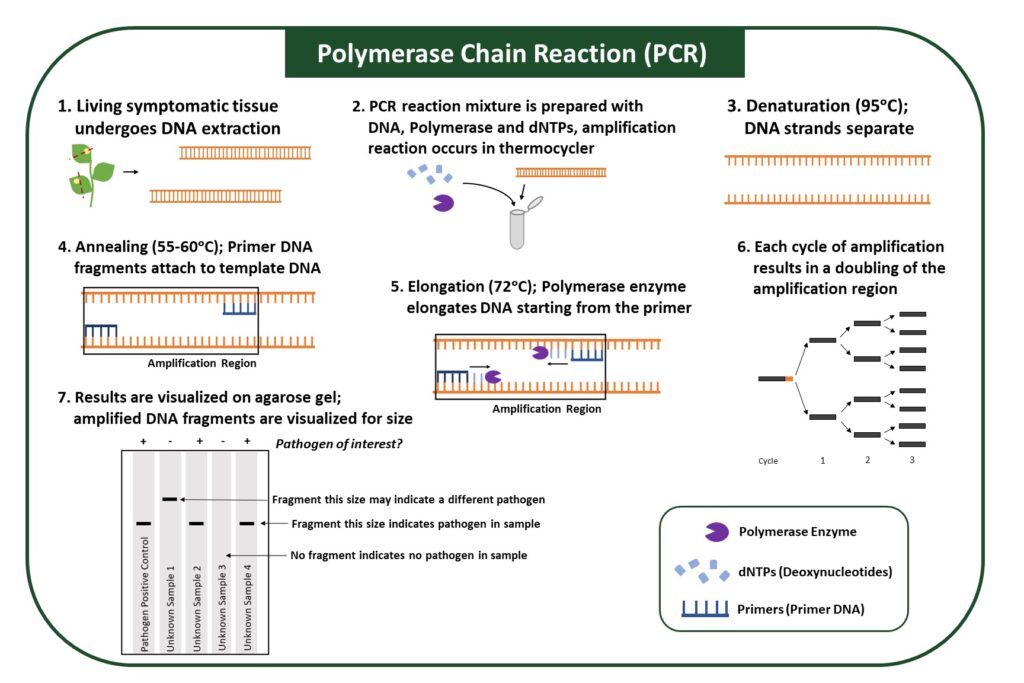
Figure 4: Polymerase Chain Reaction (PCR). PCR uses pathogen-genome-specific primer DNA and a polymerase enzyme to amplify pathogenDNA fragments. Amplified DNA fragments can be visualized with an agarose gel, where banding patterns can indicate pathogen presence in thesample.
RPA (Recombinase Polymerase Amplification)
RPA utilizes enzymes to amplify specific regions of pathogen genomes (Figure 5). Regions of amplification are designated by primers and typically designed to identify a specific pathogen. Samples that contain the pathogen will show DNA amplification, while samples that do not have the pathogen will not.
This assay is more sensitive than ELISA, or Lateral Flow Assay, because target DNA from pathogens is amplified, allowing for the detection of lower pathogen levels. This method does not require DNA extraction, and amplification steps can be performed at a broader range of temperatures. Reagents for the amplification reaction can be dried, allowing for use outside laboratories. Results can be visualized through several methods, each requiring different equipment.
RPA is a versatile assay that can be performed rapidly and with reagents that do not require special storage. This assay can easily be used with multiple samples and in the field.
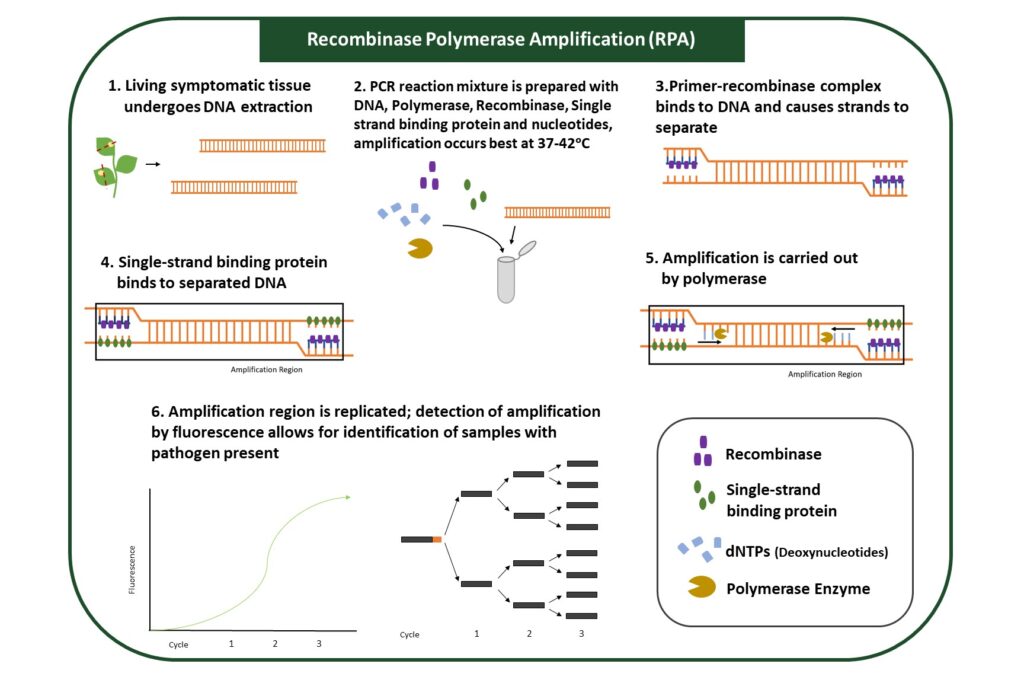
Figure 5: Recombinase Polymerase Amplification (RPA). RPA uses pathogen-genome specific primer DNA and a recombinase enzyme and polymerase enzyme to amplify pathogen DNA fragments. Amplified DNA fragments can be visualized with spectroscopy.
Next-Generation Sequencing (NGS)
NGS is a high-throughput method by which pathogens are identified based on their DNA (genomic) or RNA sequences. Other molecular methods, which target pathogen protein, or small portions of pathogen DNA, can result in false negatives if a pathogen can alter the assay target, be it protein or DNA.
Though many sequencing technologies exist, most follow a similar workflow (Figure 6). After sequencing, the generated DNA or RNA sequences can be aligned to known sequences available in public databases. This technique requires high-quality DNA or RNA extraction. Pathogen identification can be made through direct comparison to known sequences and is especially helpful for pathogens such as viruses, which can recombine quickly and be difficult to identify confidently.
In addition, sequencing allows for a comprehensive view of all pathogens in any given sample. Sequencing can be performed through different methods that provide distinct advantages that can significantly change how sequencing is performed and the complexity of computational analysis. Sequencing can be costly, and sample preparation and analysis typically require personnel with expertise.
While NGS is not currently being used in the field, technologies like Oxford Nanopore’s MinION, which is portable and relatively inexpensive, have the potential to make NGS more accessible and position it to be a helpful new tool in plant molecular diagnostics.
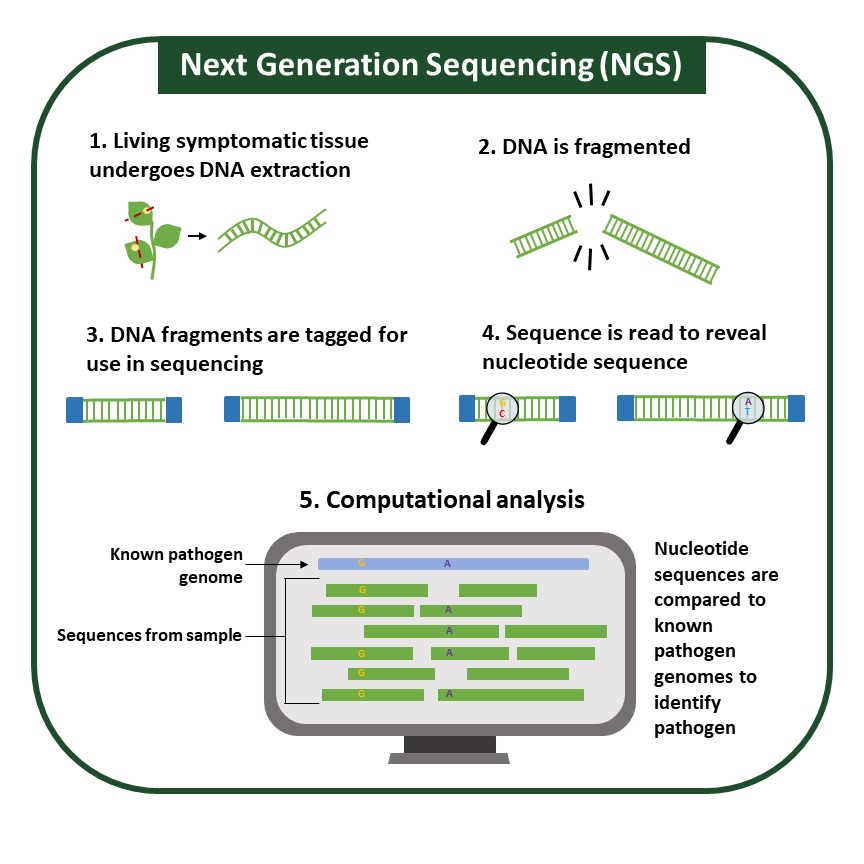
Figure 6: Sequencing Workflow. Specific sequencing technologies follow individual workflows, but in general sequencing requires the extraction of genomic material (DNA or RNA), the tagging of DNA fragments, the reading of each fragments nucleotide sequence, and then computational analysis to compare fragment sequences to known pathogen genomes.
Practical Considerations
Molecular diagnostics can identify many pathogens, but each method has practical considerations (Figure 7). A method’s sensitivity determines what concentration of pathogen a method can reliably detect. Time is also a consideration when choosing a method.
Some methods, such as RPA and Lateral Flow Assay, can be performed in a short period, typically under 30 minutes, and are therefore more appropriate for on-site and field testing. On the other hand, PCR and ELISA are more time-consuming for fewer samples but can be more efficient when more samples are being analyzed.
Molecular diagnostic techniques are likely to be more expensive than other diagnostic methods due to their requirement for equipment and materials. However, within the molecular methods, a wide range of costs are still associated with individual methods.
Finally, some methods require less expertise and equipment, such as Lateral Flow Assay and RPA, and therefore are viable options for field diagnostics. Other methods, such as ELISA, PCR, and Sequencing, require equipment or trained personnel and, therefore, typically must be performed in a laboratory.
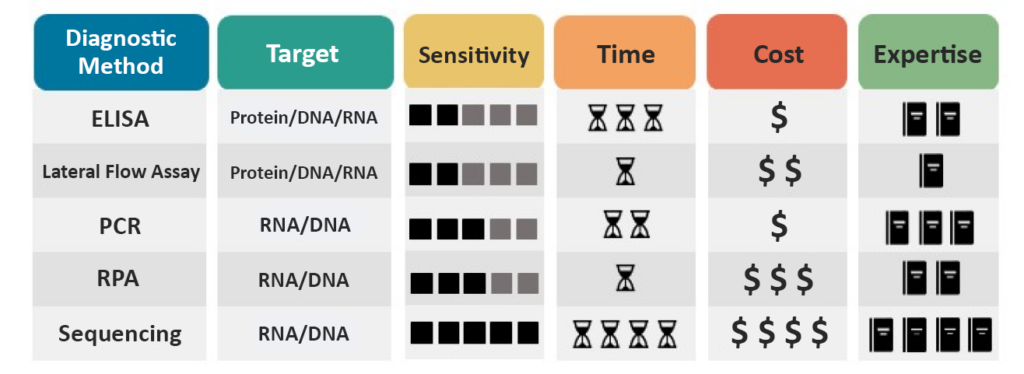
Figure 7: Practical Considerations. Each molecular plant diagnostic method has practical considerations that can be important when choosing a method for diagnostic use. These practical considerations include what the method targets, the sensitivity of the method, time, cost, and expertise.
Colorado State University Extension is an equal opportunity provider. Colorado State University does not discriminate on the basis of disability and is committed to providing reasonable accommodations. CSU’s Office of Engagement and Extension ensures meaningful access and equal opportunities to participate to individuals whose first language is not English.
Colorado State University Extension es un proveedor que ofrece igualdad de oportunidades. Colorado State University no discrimina por motivos de discapacidad y se compromete aproporcionar adaptaciones razonables. Office of Engagement and Extension de CSU garantiza acceso significativo e igualdad deoportunidades para participar a las personas quienes su primer idioma no es el inglés.
col.st/ll0t3
© Colorado State University Extension. 09/23